The NRF2-KEAP1 pathway and epigenetic control in colorectal cancer: A promising prognostic and predictive biomarker
Main Article Content
Abstract
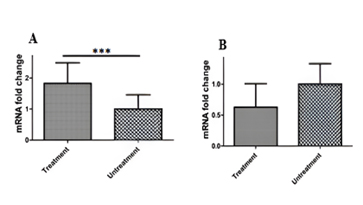
In colorectal cancer (CRC), important genes in the NRF2/KEAP1 pathway may undergo methylation, yet the exact genes that control this pathway in CRC are not entirely understood. Targeting the NRF2/KEAP1 pathway could be a valuable therapeutic strategy for colorectal cancer. Our research examined the methylation and expression of the KEAP1 gene in precancerous lesions and colorectal cancer, as well as investigated this pathway in the HT-29 cell line for potential CRC treatment. In this study, 208 frozen colorectal tissue samples were analysed, including 34 tumours, 60 precancerous lesions with adjacent normal tissues, and 20 normal tissue samples. The methylation-specific PCR and quantitative PCR were used to examine KEAP1 gene promoter methylation, and NRF2 and KEAP1 gene expressions, respectively. Furthermore, the HT 29 cell line was treated with 5-Azacytidine, followed by quantitative real-time PCR to evaluate post-demethylation KEAP1 and NRF2 mRNA expression. Unlike NRF2, our research revealed a notable reduction in KEAP1 gene expression in CRC tissues compared to adjacent normal tissues (p=0.02). Intriguingly, treating the HT-29 cell line with 5-Azacytidine led to a significant increase in KEAP1 gene expression compared to untreated cells (P=0.03), with no significant alteration observed in NRF2 gene expression. Examination of CpG islands within the KEAP1 gene promoter indicated extensive hypermethylation in 58.8% of tumour tissues compared to adjacent normal samples. These findings indicate that decreased KEAP1 gene expression resulting from promoter hypermethylation could interfere with the NRF2/KEAP1 pathway, a crucial mechanism in colorectal carcinogenesis. The study proposes that KEAP1 may play a significant role in the progression of CRC and could potentially serve as a biomarker for diagnosing and monitoring therapy in CRC
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.